Setup packrat project for working on remote databases with R
Overview
As a data analyst I often consult for clients who need me to work on their existing infrastructure. But this requires me to set up my entire environment on a remote machine.
So the question is; “What is the best way for me to setup an environment for database exploration on a clean machine?”
Introducing packrat - R’s package management system.
Installation
The first thing you want to do is setup the environment you want to use on your local machine. This is important when the machine you will be given access to is entirely isolated from the internet (strict firewall setup).
- Install packrat:
install.packages("packrat")You need to install packrat first before you can create a version controlled repository for your project.
- Create a new packrat project:
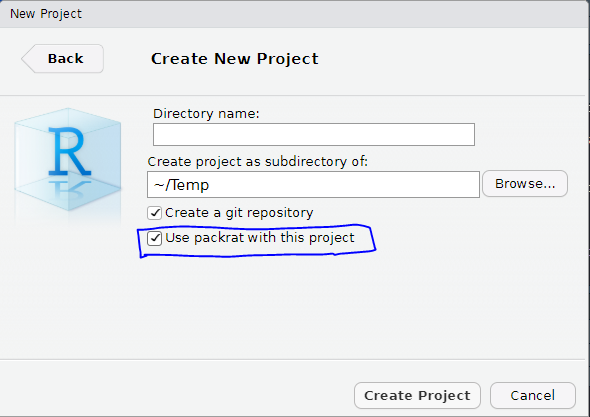
The packrat project is completely isolated from your usual R libraries. In fact it uses the libraries inside the project folder at packrat/lib-R
In my case R tried to load the EightyR package on startup but couldn’t find it:
- Install essential packages
- install.packages(“tidyverse”)
- install.packages(“odbc”)
- install.packages(“DBI”)
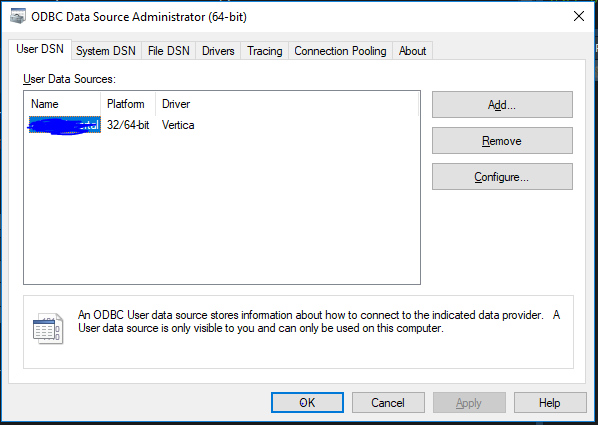
- Make sure the ODBC connections are setup on the client side machine for the databases you are interested in.
For example if you are working on a Windows machine you will have to use the ODBC wizzard to setup all the drivers;
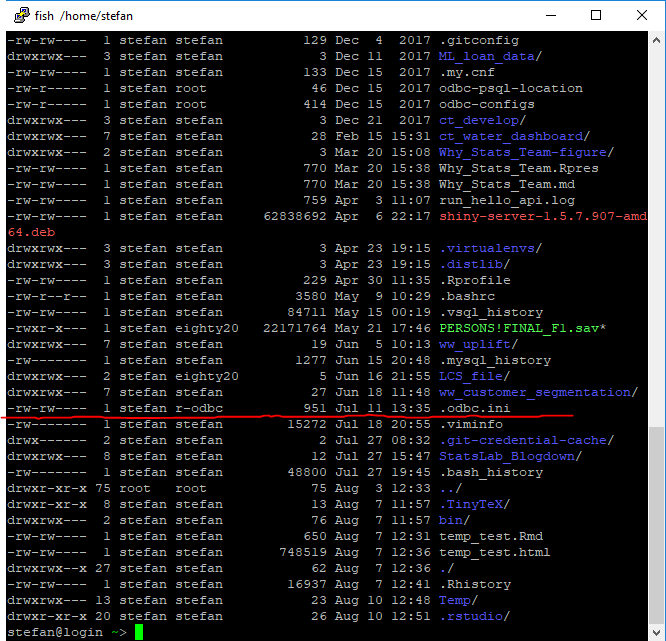
But if you are on a linux machine you will need to setup the .odbc.ini file in your home directory (the one you will be using R-studio on)
My file can be found like this:
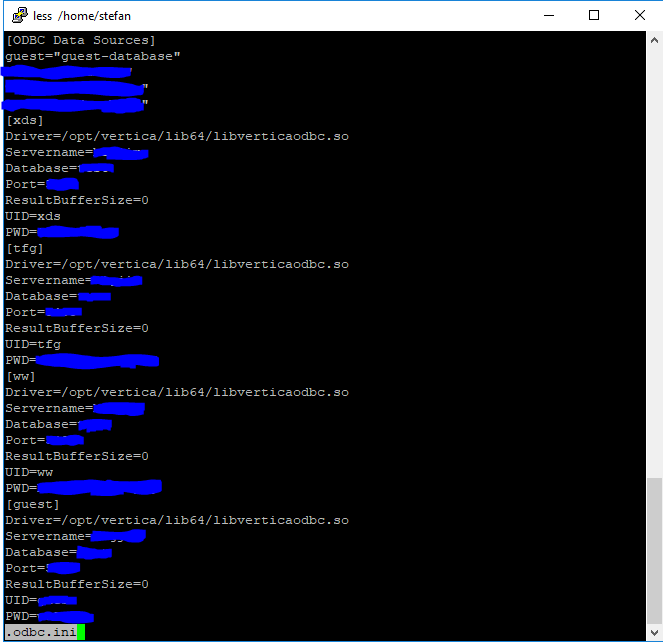
setup your file to match the database drivers you are using, for example:
- Test your connections tab in R-studio
The connections tab makes our lives easier by picking up all the drivers and connections we have installed on the running machine.
We can therefore use the connections tab to open a connection to the databases and then run some analytics on them.
Your connections tab should look something like this:
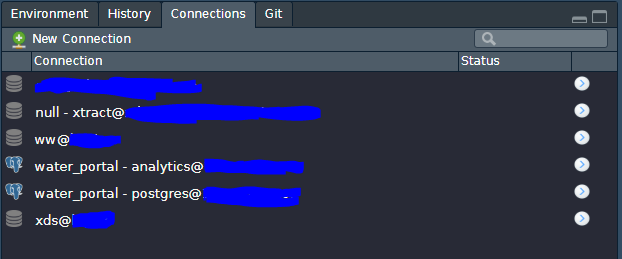
Click on the database you want to test and copy the connection string to your Rmd or script:
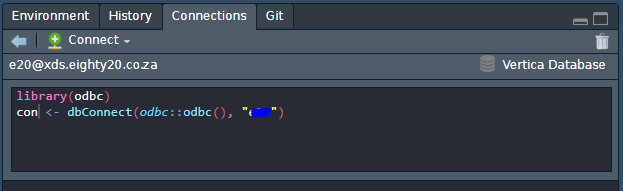
Make sure that you can connect to the database:
translation_con <- dbConnect(odbc::odbc(), "e20_translation")
example_tbl <- tbl(translation_con, sql("select * from eighty20_databaseadmin_uat.translation_db"))
example_tbl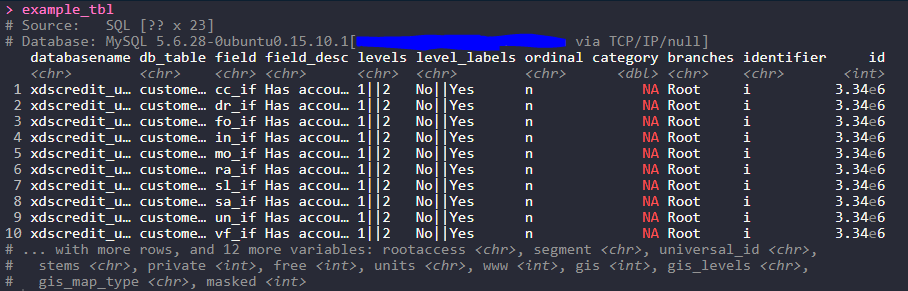
dbplyr
Do we have to write sql for every query??
You can simply write your query in the sql() expression. But this is seldom done because we can leverage dbplyr.
What you need to know;
- dbplyr syntax works exactly like dplyr syntax
- dbplyr code gets executed in database by using sql translation
- dbplyr code prints the output to the screen by executing the code in database
This means your output never touches your R environment unless you tell it to!
Example of temporary data
Let’s execute a dbplyr chain and save it to an object called temporary_tbl:
temporary_tbl <-
example_tbl %>%
group_by(ordinal) %>%
tally
temporary_tbl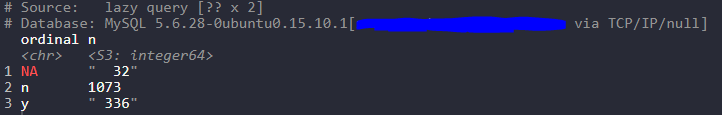
In standard dplyr fashion we have counted the number of rows describing wether or not a field is a nominal field.
It is tempting then to assume that this data in fact lives in the variable temporary_tbl. This is not the case:
temporary_tbl %>% str
What is this voodoo!?
When we call the variable temporary_tbl we are actually executing the query in database and then redirecting the output of the terminal to our R console.
collect() statement
If we wanted to further use this in a ggplot for example we need to call the collect() statement:
in_memory <- example_tbl %>%
group_by(ordinal) %>%
tally %>%
collect
in_memory %>% class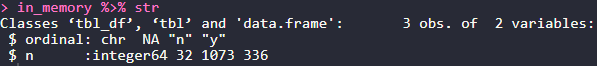
We see that the collect statement creates a tibble object for us. Unfortunately most databases will do numeric calculations in a format integer 64 for precision. Although this is a good thing the dbplyr package does not type cast this for us explicitly to a numeric type variable…
Leaving this data type as integer 64 will cause many headaches later so it’s a good idea to create a wrapper that can correct this for us:
convert_int_64 <- function(df){
is_int_64 <- function(x){
ifelse(class(x)=="integer64",T,F)
}
return(df %>% mutate_if(is_int_64,as.numeric))
}We can execute this chain to plot the table:
example_tbl %>%
group_by(ordinal) %>%
tally %>%
collect %>%
convert_int_64 %>%
ggplot()+
geom_bar(stat='identity', aes(x=ordinal,y=n, fill = ordinal))+
ggtitle("Collected dbplyr chain and then plotted a bar chart")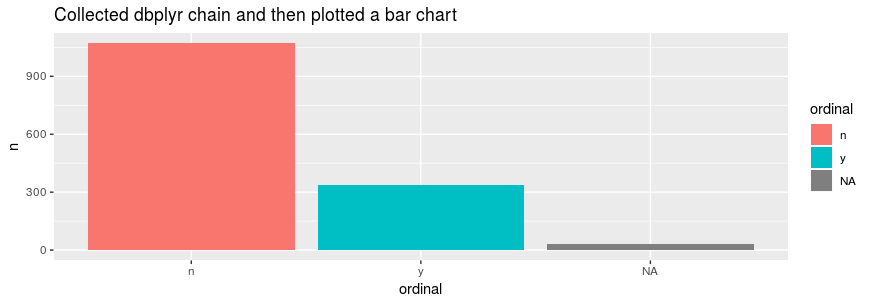
When dbplyr cannot translate your chain into a lazy query you can do one of the following:
- Call the collect() statement earlier in the chain and then carry on in memory
- Write a file_name.sql file and call it using example_tbl <- tbl(translation_con, sql(read_file("file_name.sql")))
Compile your project
Now that you have tested all the tools and packages you will need you can commit all your work and push it to a remote repository for version control.
When moving over to the new machine make sure that the IT team on the clients side can clone the repository into your R-studio or user home directory.
Launching the packrat project in this folder will automatically load up all the packages and versions you installed on your local machine
